-Search query
-Search result
Showing 1 - 50 of 564 items for (author: wu & gh)

EMDB-41363: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5
Method: single particle / : Yue H, Hunkeler M, Roy Burman SS, Fischer ES

PDB-8tl6: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5
Method: single particle / : Yue H, Hunkeler M, Roy Burman SS, Fischer ES

EMDB-33347: 
Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state)
Method: single particle / : Wu Z, Yu Z, Tan S, Lu J, Lu G, Lin J

PDB-7xog: 
Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state)
Method: single particle / : Wu Z, Yu Z, Tan S, Lu J, Lu G, Lin J

EMDB-41625: 
Type IV pilus from Pseudomonas PAO1 strain
Method: single particle / : Thongchol J, Zhang J, Zeng L

EMDB-41632: 
Asymmetric reconstruction of mature PP7 virions
Method: single particle / : Thongchol J, Zhang J, Zeng L

EMDB-41633: 
Type IV pilus from Pseudomonas PAO1 strain with PP7 Maturation protein
Method: single particle / : Thongchol J, Zhang J, Zeng L

EMDB-41675: 
The original consensus map of PP7 binding to T4P from PAO1
Method: single particle / : Thongchol J, Zhang J, Zeng L

EMDB-41780: 
Composite map of PP7 binding to type IV pilus from PAO1
Method: single particle / : Thongchol J, Zhang J, Zeng L

PDB-8tum: 
Type IV pilus from Pseudomonas PAO1 strain
Method: single particle / : Thongchol J, Zhang J, Zeng L

PDB-8tuw: 
Type IV pilus from Pseudomonas PAO1 strain with PP7 Maturation protein
Method: single particle / : Thongchol J, Zhang J, Zeng L

PDB-8tux: 
Capsid of mature PP7 virion with 3'end region of PP7 genomic RNA
Method: single particle / : Thongchol J, Zhang J, Zeng L

EMDB-33346: 
Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state)
Method: single particle / : Wu Z, Yu Z, Tan S, Lu J, Lu G, Lin J

PDB-7xoe: 
Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state)
Method: single particle / : Wu Z, Yu Z, Tan S, Lu J, Lu G, Lin J

EMDB-41374: 
Antibody N3-1 bound to RBDs in the up and down conformations
Method: single particle / : Hsieh CL, McLellan JS
EMDB-41382: 
Antibody N3-1 bound to RBD in the up conformation
Method: single particle / : Hsieh CL, McLellan JS
EMDB-41399: 
Antibody N3-1 bound to SARS-CoV-2 spike
Method: single particle / : Hsieh CL, McLellan JS

PDB-8tm1: 
Antibody N3-1 bound to RBDs in the up and down conformations
Method: single particle / : Hsieh CL, McLellan JS

PDB-8tma: 
Antibody N3-1 bound to RBD in the up conformation
Method: single particle / : Hsieh CL, McLellan JS
EMDB-35463: 
Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state)
Method: single particle / : Pan X, Fang Y
EMDB-35483: 
Cryo-EM structure of human HCAR2-Gi complex with niacin
Method: single particle / : Pan X, Fang Y
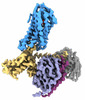
EMDB-35484: 
Cryo-EM structure of human HCAR2-Gi complex with acipimox
Method: single particle / : Pan X, Fang Y
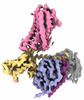
EMDB-35485: 
Cryo-EM structure of human HCAR2-Gi complex with MK-6892
Method: single particle / : Pan X, Fang Y

PDB-8ij3: 
Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state)
Method: single particle / : Pan X, Fang Y

PDB-8ija: 
Cryo-EM structure of human HCAR2-Gi complex with niacin
Method: single particle / : Pan X, Fang Y

PDB-8ijb: 
Cryo-EM structure of human HCAR2-Gi complex with acipimox
Method: single particle / : Pan X, Fang Y

PDB-8ijd: 
Cryo-EM structure of human HCAR2-Gi complex with MK-6892
Method: single particle / : Pan X, Fang Y

EMDB-36076: 
Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2
Method: single particle / : Yang GH, Zhang YM, Zhou JQ, Jia YT, Xu X, Fu P, Wu HY

EMDB-36077: 
Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1
Method: single particle / : Yang GH, Zhang YM, Zhou JQ, Jia YT, Xu X, Fu P, Wu HY

PDB-8jd9: 
Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1
Method: single particle / : Yang GH, Zhang YM, Zhou JQ, Jia YT, Xu X, Fu P, Wu HY

PDB-8jda: 
Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2
Method: single particle / : Yang GH, Zhang YM, Zhou JQ, Jia YT, Xu X, Fu P, Wu HY

EMDB-36326: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist peptide15-bound human GLP-1R-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

PDB-8jis: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist peptide15-bound human GLP-1R-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

EMDB-35155: 
Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab
Method: single particle / : Li Z, Yu F, Cao S, ZHao H

EMDB-35156: 
Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab
Method: single particle / : Li Z, Yu F, Cao S

PDB-8i3s: 
Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab
Method: single particle / : Li Z, Yu F, Cao S, ZHao H

PDB-8i3u: 
Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab
Method: single particle / : Li Z, Yu F, Cao S

EMDB-36324: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist Peptide 15-bound human GCGR-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

EMDB-36328: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GCGR-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

PDB-8jiq: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist Peptide 15-bound human GCGR-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

PDB-8jiu: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GCGR-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

EMDB-36323: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GLP-1R-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

EMDB-36325: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GLP-1R-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

EMDB-36327: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GCGR-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

PDB-8jip: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GLP-1R-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

PDB-8jir: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GLP-1R-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

PDB-8jit: 
Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GCGR-Gs complex
Method: single particle / : Yang L, Zhou QT, Dai AT, Zhao FH, Chang RL, Ying TL, Wu BL, Yang DH, Wang MW, Cong ZT

EMDB-36474: 
cryo-EM structure of NTSR1-GRK2-Galpha(q) complexes 1
Method: single particle / : Duan J, Liu H, Zhao F, Yuan Q, Ji Y, Xu HE

EMDB-36475: 
cryo-EM structure of NTSR1-GRK2-Galpha(q) complexes 2
Method: single particle / : Duan J, Liu H, Zhao F, Yuan Q, Ji Y, Xu HE
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
